Research
Welcome to the Schärfen Lab at the RNA Therapeutics Institute (RTI) of UMass Chan. Right now, we are only beginning to recognize the important role of RNA structure in molecular and cellular biology. Our lab studies RNA structure from a global, systems-level perspective: through large-scale comparative measurement of structure, and through systematic functional characterization. Most of our experiments are designed to answer questions about RNA structure in vivo. Our approaches draw from fields including genomics, data science, systems biology, biochemistry, and others.
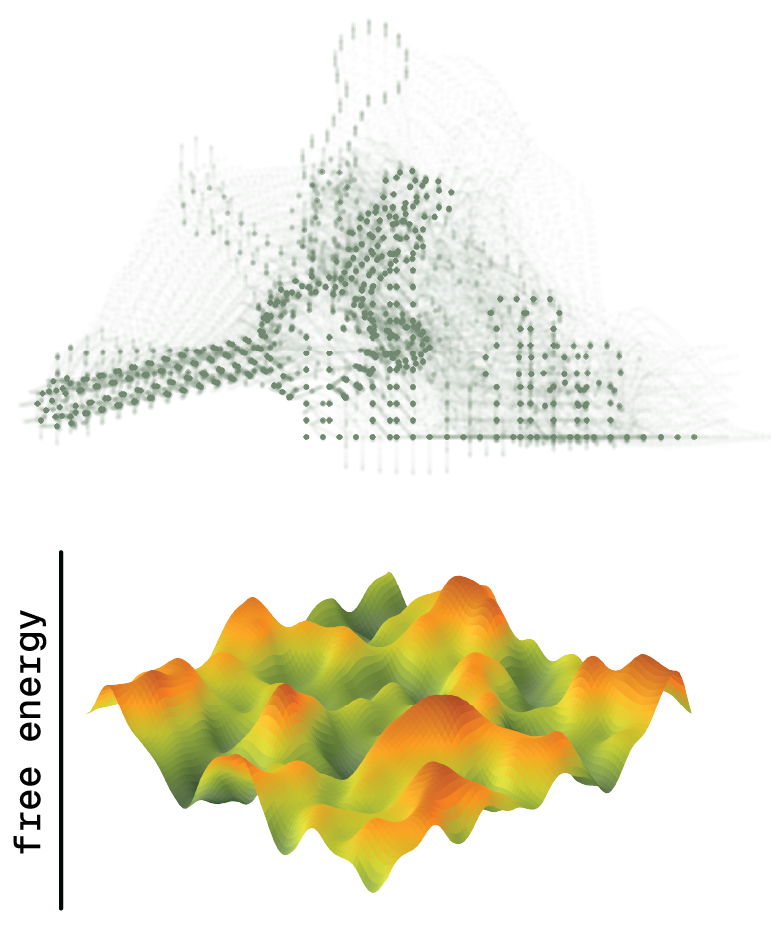
RNA structure is dynamic
Each individual RNA molecule can adopt many distinct conformations, which differ in their stability, or free energy of folding (∆G). Each RNA sequence therefore defines a unique free energy landscape, where stable structures populate the valleys. Unlike for proteins, RNA folding landscapes are typically rugged, such that RNAs may exist in several equally stable conformations. We are interested in how this unique property shapes the molecular functions of natural and synthetic RNAs.
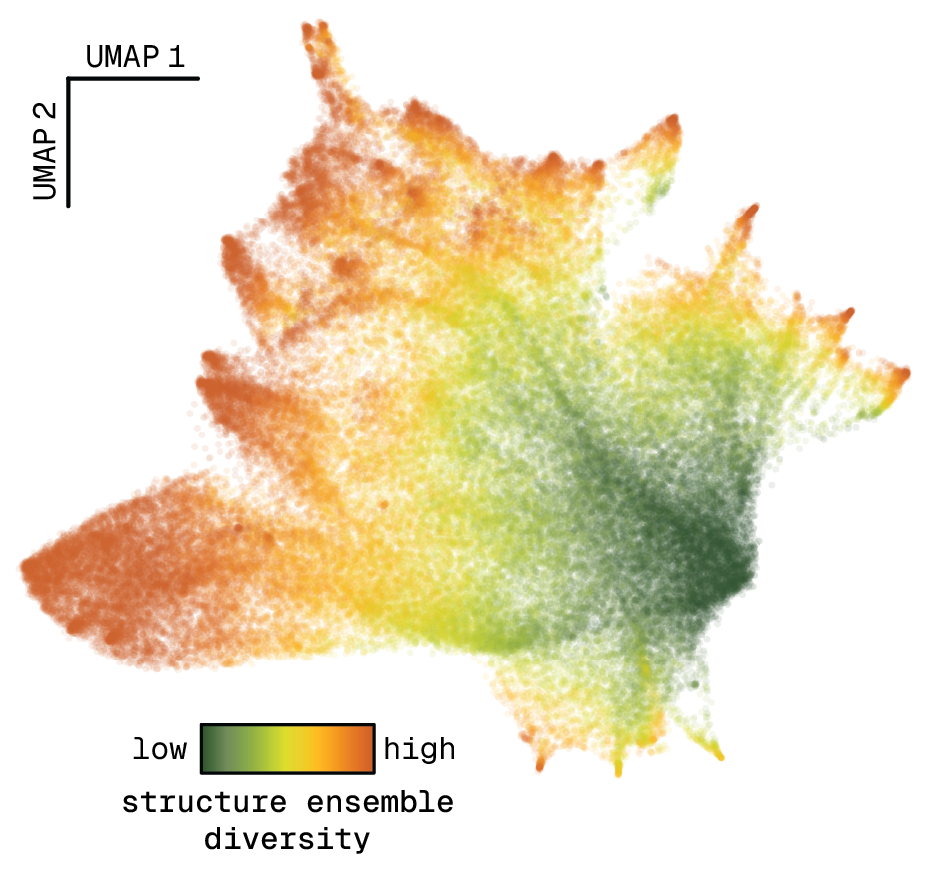
Large-scale data collection
Natural RNAs explore a huge sequence space, and because they are dynamic, the structure space is even larger. We aim to measure RNA structure and function of many thousands of sequences. To do this, we use different sequencing methods and massively parallel reporter assays. Then, we apply machine learning methods to extract information from the large-scale data sets that we collect in the lab.

Structure variants
To make sense of the structural features that confer an RNA's function, we construct RNA libraries where each sequence represents a slightly different structure. By characterizing structurally similar RNA variants, we can map structure to function at high resolution, directly inside cells.
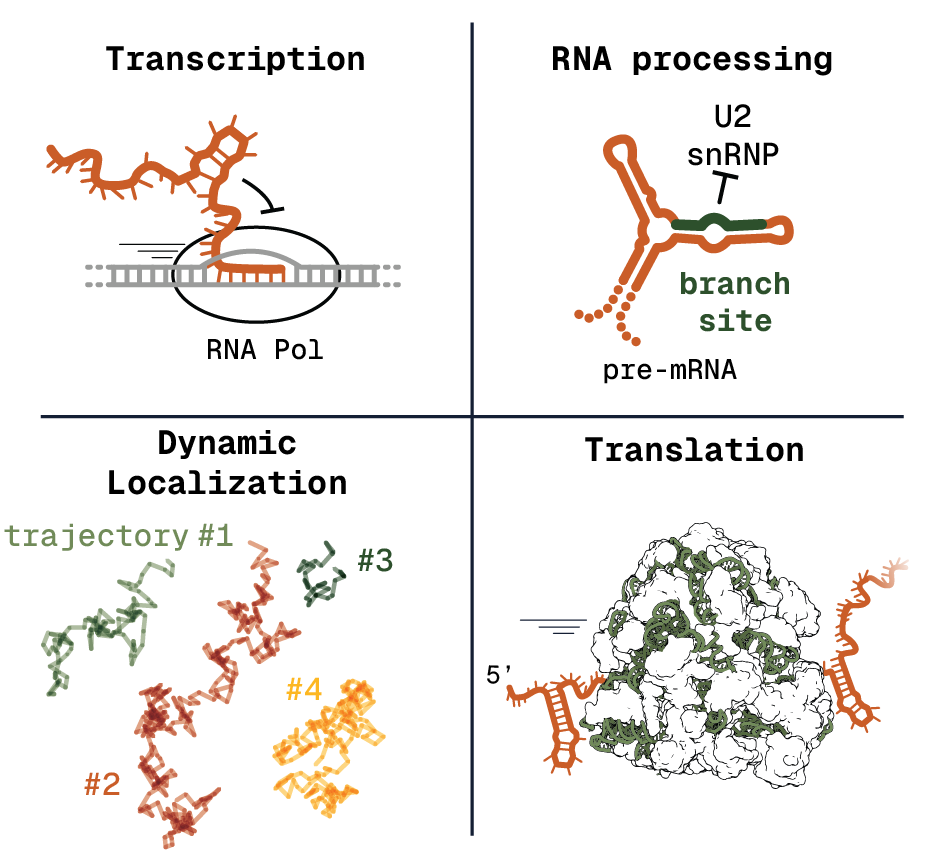
Molecular Processes
Formation of RNA structure has been shown to influence many cellular processes. We aim to obtain a comprehensive overview of the mechanisms and effect sizes at each step of gene expression. This includes transcription, pre-mRNA splicing, cellular localization and translation.
Methods
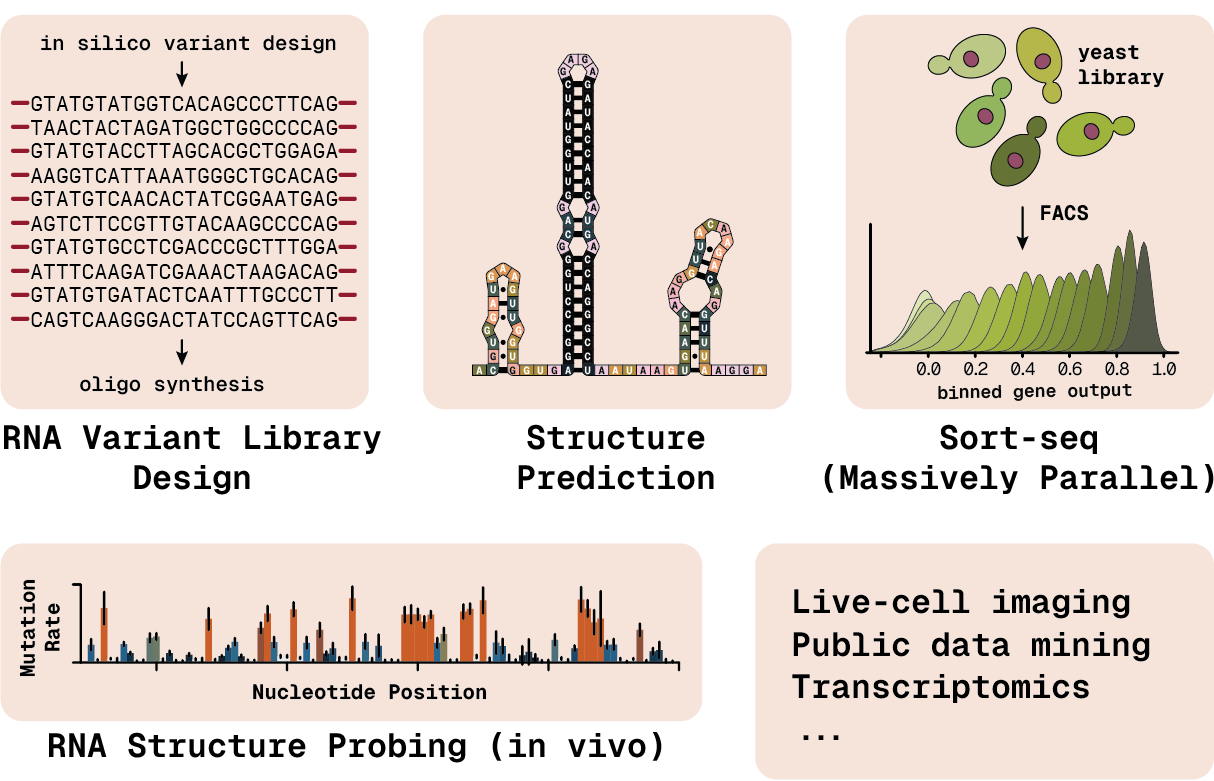
Our lab applies a range of methods to produce and analyze large data sets. We use RNA structure prediction algorithms to design variant libraries that we can test in cells. Massively parallel reporter assays such as Sort-seq allow us to assign function to tens of thousands of sequences in a single experiment. We directly probe RNA base pairing or structural flexibility inside cells with tools like DMS-MaPseq, CoSTseq, and SHAPE-based methods. We always look to apply or develop new methods to add to our repertoire!